Imaging and Electrode Localization #
Description #
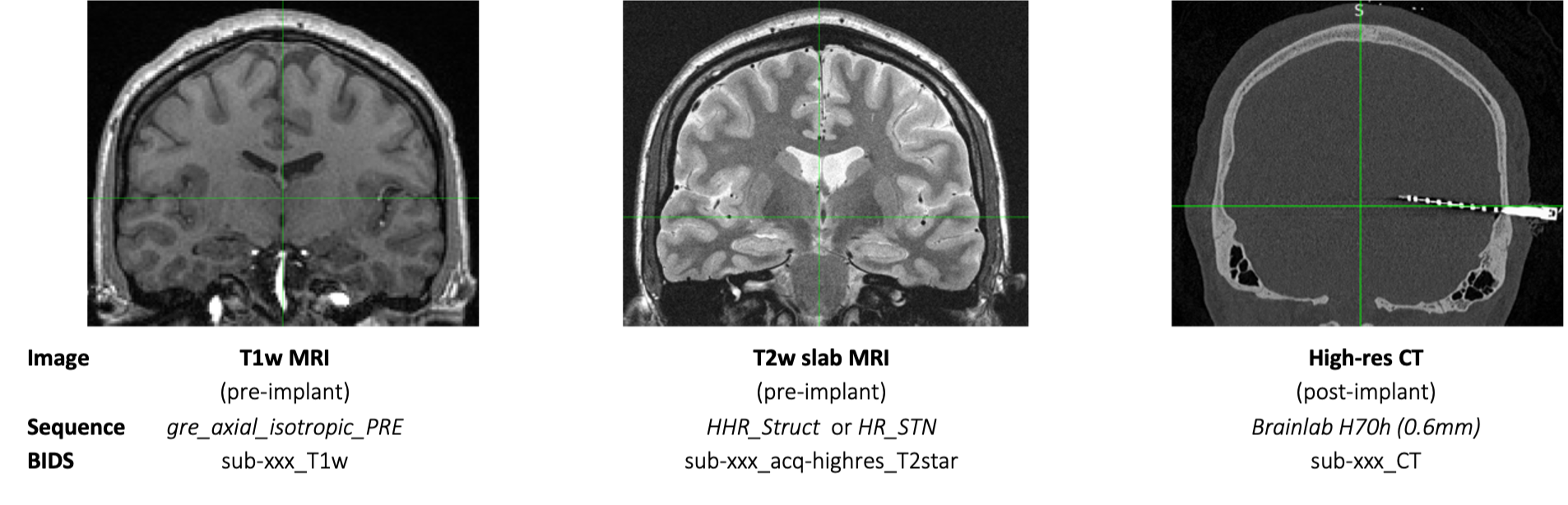
Implanted depth electrodes are localized by aligning the pre-implant T1w MRI (1mm isotropic) and post-implant high-resolution CT (0.6mm), which shows electrode contacts as hyperintensities. An additional pre-implant image, a high-res T2w slab MRI, provides a high-resolution image of medial temporal lobe regions. The T1w and T2w slab images are used to identify the underlying anatomy at each electrode contact location. Spatial normalization to MNI 152 template brain can be used for comparison of regions across groups. Automated image processing algorithms such as ASHS and FreeSurfer are implemented to quickly and reliably segment anatomical structures to aid in localization. Imaging data are structured according to BIDS specifications.
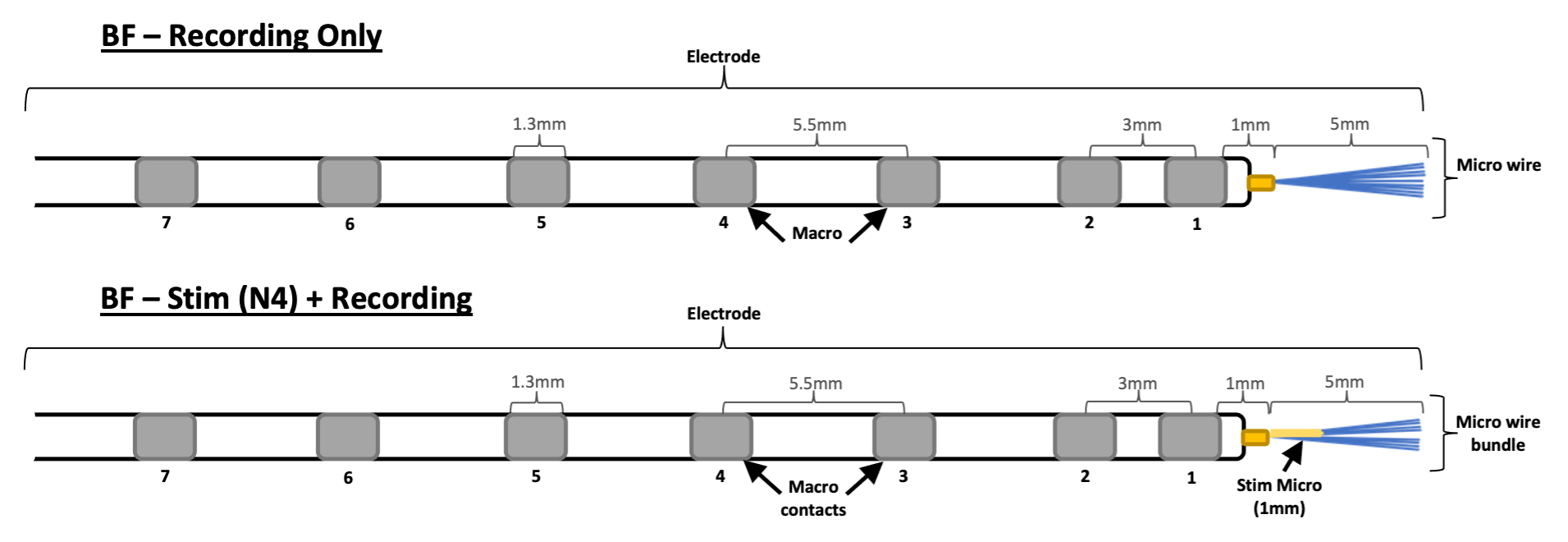
Electrode Specifications #
Behnke-Fried (BF) #
- macros and micros
- two fabrications:
1. In-house micros (fabricated by Chris Chan, Clinical Specialist)
2. Commercial micros (purchased from Ad-Tech Medical Instrument Corp.)
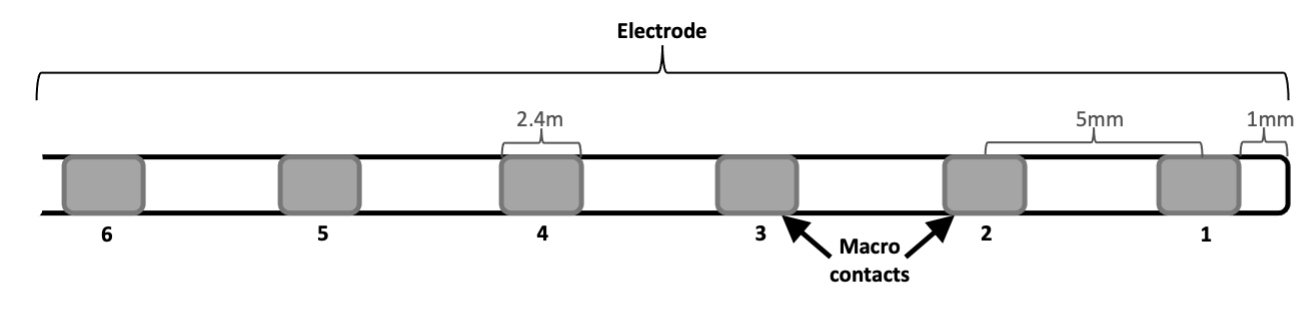
Spencer Depth (SD) #
- macros only
Download DICOMs #
Brainlab Elements (Origin Server) #
Go to vspbrainlbap01.ad.medctr.ucla.edu
- Enter Mednet credentials
- Click “Select Patient”
- Enter patient’s MRN in search bar and select the correct patient
- Verify MRN, name, and DOB before proceeding
- Click “Viewing”
- Search for images (scrolling leftwards “back in time”)
- Look for dates for pre-op MRI (day of consent) and post-op CT (day of surgery)
- Images to download, find corresponding sequences:
Image
Sequence
BIDS suffix
T1w MRI (pre-implant)
gre_axial_isotropic_PRE
_T1w
T2w Slab MRI (pre-implant)
HHR_Struct or HR_STN
_acq-highres_T2star
High-Res CT (post-implant)
Brainlab H70h (0.6mm)
_CT
- Select one image at a time:
- View image to verify, then click “Home” icon
- Click “Export”
- Select “Brainwash” (anonymizes images), then “Download to Client” (downloads to browser)
- Move to CNL server
- /10.44.239.160/data/ImagingData/BIDS/sourcedata/sub-[sub]/[anat,ct]
Electrode Localization Pipeline #
Convert DICOMs to BIDS #
- img2bids_lts.sh
- location: […]/ImagingData/BIDS/code/scripts
- converts DICOMs to NIfTI and BIDS-compliant images
- fs_batch.sh
- location: […]/ImagingData/BIDS/code/freesurfer_7.3.2
- standard recon-all plus two hippoamygdalar segmenation algorithms
Run LeGUI #
- runLeGUI.sh
- see LeGUI installation
- launches LeGUI (see LeGUI protocol)
LeGUI to .xlsx and Hoffman PDM .mat #
- mkLeGuiExcel.m
- location: […]/LTS/ImagingData/BIDS/code/legui
- makes Excel table from LeGUI output and saves to subject’s BIDS/derivatives/legui/… directory on LTS
- generateLeGuiSummary.m
- location: […]/Hoffman/data/PIPELINE_vc/CODE/ImagingAndAnatomy
- makes Matlab structure identical to LeGUI Excel table and saves to subject’s Patient Database directory on Hoffman
Convert LeGUI to BIDS #
- legui2bids_v3.py
- location: […]/ImagingData/BIDS/code/legui
- converts LeGUI output into BIDS-compliant table (.tsv) and saves to BIDS/sub-[sub]/ieeg
- ashs_prep_t1_t2_parallel.sh
- location: […]/ImagingData/BIDS/code/ashs/itksnap_cloud
- prepares T1w and T2w slab for ASHS submission
Run ASHS #
- ITKSNAP CLOUD (DSS) ASHS
- launch ITKSNAP 4.0 and submit images to DSS with ASHS ABC (Berron ’17)
- download results to […]/BIDS/derivatives/ashs/itksnap_cloud/sub-[sub]/ashs_abc
Generate Coordinates and Images #
- elec_post_proc.py
- location: […]/ImagingData/BIDS/code/ashs/itksnap_cloud
- transforms micro and macro coords from LeGUI into different image spaces and ultimately makes an image of electrode markers (crosshairs) for ASHS and FreeSurfer
Coregistration QC #
ASHS T2w – LeGUI T1w images #
- Check coregistration between the images at […]/derivatives/ashs/ashs_prep/sub-xxx
- sub-xxx_desc-ashs_T2star.nii.gz
- ASHS T2w hippocampal slab
- sub-xxx_desc-legui_T1w.nii.gz
- LeGUI T1w
- sub-xxx_space-ashsT2_desc-legui_T1w.nii.gz
- LeGUI T1w coregistered to FreeSurfer T1w-space
- sub-xxx_desc-ashs_T2star.nii.gz
- If sub-xxx_desc-ashs_T2star.nii.gz and sub-xxx_space-ashsT2_desc-legui_T1w.nii.gz are not coregistered properly, then complete the following:
- launch ITKSNAP 4.0 with sub-xxx_desc-ashs_T2star.nii.gz (main image) and sub-xxx_desc-legui_T1w.nii.gz (additional image)
- Use Registration tool (cmd + R) with these Automatic tab settings:
- moving image layer = sub-xxx_desc-legui_T1w.nii.gz
- transformation model = rigid
- image similarity metric = mutual information
- multi-resolution schedule:
- coarsest level = 8x
- finest level = 4x
- Click Run Registration
- Check registration at multiple landmarks in all three slice views
- Save transformation matrix (button at bottom) as ITK “.txt” file
- Convert ITK “.txt” to FSL “.mat” in Terminal:
- c3d_affine_tool -ref sub-xxx_desc-ashs_T2star.nii.gz -src sub-xxx_desc-legui_T1w.nii.gz -itk sub-xxx_space-ashsT2_desc-itksnap-legui_T1w.txt -ras2fsl -o sub-xxx_space-ashsT2_desc-legui_T1w.mat
- c3d_affine_tool -ref sub-xxx_desc-ashs_T2star.nii.gz -src sub-xxx_desc-legui_T1w.nii.gz -itk sub-xxx_space-ashsT2_desc-itksnap-legui_T1w.txt -ras2fsl -o sub-xxx_space-ashsT2_desc-legui_T1w.mat
- Re-run elec_post_proc.py
FreeSurfer T1w – LeGUI T1w images #
- Check coregistration between the images at […]/derivatives/freesurfer_7.3.2/sub-xxx/elec_loc
- sub-xxx_desc-FS_T1w.nii.gz
- FreeSurfer T1w
- sub-xxx_desc-legui_T1w.nii.gz
- LeGUI T1w
- sub-xxx_space-FS_desc-legui_T1w.nii.gz
- LeGUI T1w coregistered to FreeSurfer T1w-space
- sub-xxx_desc-FS_T1w.nii.gz
- If sub-xxx_desc-FS_T1w.nii.gz and sub-xxx_space-FS_desc-legui_T1w.nii.gz are not coregistered properly, then complete the following:
- launch ITKSNAP 4.0 with sub-xxx_desc-FS_T1w.nii.gz (main image) and sub-xxx_desc-legui_T1w.nii.gz (additional image)
- Use Registration tool (cmd + R) with these Automatic tab settings:
- moving image layer = sub-xxx_desc-legui_T1w.nii.gz
- transformation model = rigid
- image similarity metric = mutual information
- multi-resolution schedule:
- coarsest level = 8x
- finest level = 4x
- Click Run Registration
- Check registration at multiple landmarks in all three slice views
- Save transformation matrix (button at bottom) as ITK “.txt” file
- Convert ITK “.txt” to FSL “.mat” in Terminal:
- c3d_affine_tool -ref sub-xxx_desc-FS_T1w.nii.gz -src sub-xxx_desc-legui_T1w.nii.gz -itk sub-xxx_space-FS_desc-itksnap-legui_T1w.txt -ras2fsl -o sub-xxx_space-FS_desc-legui_T1w.mat
- c3d_affine_tool -ref sub-xxx_desc-FS_T1w.nii.gz -src sub-xxx_desc-legui_T1w.nii.gz -itk sub-xxx_space-FS_desc-itksnap-legui_T1w.txt -ras2fsl -o sub-xxx_space-FS_desc-legui_T1w.mat
- Re-run elec_post_proc.py
Capture ASHS Localizations #
- elec_ashs_review.sh
- location: […]/ImagingData/BIDS/code/ashs/itksnap_cloud
- opens subject’s micro coords and LeGUI Excel table, then launches FSLeyes with acq-highres_T2star, ASHS label image, and electrode crosshair image
- add columns to Excel table: “ASHS_ABC” and “ashs_border”
- fill with ASHS label and neighboring label if there is a shared edge or vertex
- enter “NaN” for no label
Capture FreeSurfer Localizations #
- append_coords_v3.py
- location: […]/ImagingData/BIDS/code/pipelines/cnl_elec_loc/modules
- extracts FreeSurfer regions and appends the localizations spreadsheet
Update Patient Database on Hoffman #
- update_PDM_loc_table.py
- location: […]/ImagingData/BIDS/code/pipelines/cnl_elec_loc/modules
- updates electrodeLocalizations.mat with final localizations table fields
- saves a localizations.xlsx to Hoffman Patient Database
