NOTE: our pipeline is currently a work in progress. Email jsakon–at–mednet.ucla.edu for questions.
The below information is for our unpacked data which is technically just an interim step to what will ultimately be a NWB file structure containing the data of the full experimental series and all associated metadata within one file.
The purpose of this document is for people to be able to use our data in the meantime.
The below is currently true for participants 551 onwards
Data extraction code #
The code used for data extraction can be found here. It contains a README that has additional information to that detailed below.
Glossary #
Experimental series: all task epochs run by the patient as part of the 24 movie task.
Task epoch: a specific task within the experiment eg “movie watch” or “free recall” or “sleep” etc
Experiment: here we define experiment based on the folder structure of saved data. Some experiment folders contain more than one task epoch. Experiment folders typically contain a single Neuralynx (NLX) or BlackRock (BRK) data folder.

NLX data folder: YYYY-MM-DD_hh-mm-ss folder created to store recorded data (can contain many recordings).
Recording: a group of files that share the same suffix (i.e. no suffix, _0001, _0002). Additional suffixes are created when NLX recordings are paused or automatically after 2 hours of continuous recording.
Data stream: a specific type of saved data, eg macro, micro, events, audio etc
fs/SR: sample rate
ts: timestamp, time associated with a single event (eg one neural data point, a TTL event)
index: (of time series) ordinal position within sequence
Unix time: timestamps that represent the number of seconds that have elapsed since January 1, 1970, at 00:00:00 UTC
unpacking/extraction: the process of reading a raw neural data file (eg .ncs), maybe doing some processing to get the correct units, generating timestamps, and saving in a format to be used for further analysis
DAQ: data acquisition, typically referring to the Neuralynx or BlackRock neural recordings/devices.
MovieParadigm task epochs #
The task epochs are typically run in the following order:
- Non-24 video watch
- Non-24 free recall
- 24 video watch
- 24 free recall
- 24 recognition memory task
- 24 cued recall
- Sleep
- 24 free recall
- 24 recognition memory task
- 24 cued recall
- 24 concept fixation
- Non-24 free recall
Not all participants have done the full run.
Non-24 movies were introduced at P566.
Brief description of the memory tasks #
Free recall: participant asked to verbally recall everything they can about the episode just watched.
Recognition memory: participant shown a series of short clips from either the video they just watched (targets) or a different video (foils i.e different episode of the same TV show). After each clip participant has to respond with a mouse click whether the clip is a target or foil.
Cued recall: participant shown a question with an associated image which they must verbally respond to.
Concept fixation: participant shown an image of a concept that they must hold in memory.
Schematic of recordings in a typical experiment series #

Presleep includes movie watch + memory tasks. It can contain multiple experiments.
Sleep recordings contain only one experiment but will always have multiple recordings.
Postsleep includes only memory tasks. It can also contain multiple experiments.
The *_000X.ncs extensions kick in after 2 hours of recording, where NLX will start a new recording with a different suffix. Note that (unfortunately) NLX will not necessarily record the *_000X.ncs files in logical order. E.g. *_0002.ncs might come before *_0001.ncs. This is dealt with in the extraction to unpacked mat files but just a warning if you are dealing with raw recordings.
Reference time frames #
There are a number of different reference time frames that could be used for these data.
- Global Time Frame (Unix Time Reference): The Neuralynx timestamps are recorded in Unix time. Every event or recording is placed in the continuous, system-wide timeline, starting from the Unix epoch (January 1, 1970). This provides a universal reference point across all recordings.
- Relative Time Frames : This refers to time measured relative to another timestamp. All subsequent timestamps are adjusted to start from 0 at that first reference timestamp, essentially “resetting” time to a local reference.
There are multiple relative time frames we could use:
- Series reference time frame: This time frame refers to time measured relative to the first timestamp of the first recording of the first experiment in the experimental series.
- Individual experiment time frame (per-experiment reference): The timestamps for each recording is referenced relative to the first timestamp of that specific experiment.
- Individual recording time frame (per-recording reference): The timestamps for each recording is referenced relative to the first timestamp of that specific recording.
Within all the single experiment folders everything is in Unix time.
In the cross-experiment folders, the spike times are in the series time frame. The Unix time used as reference (aka t0_unix) for the series time frame can be found in “CSC_micro_spikes_meta.json” files. Any event unix time minus t0_unix will be aligned to the spikes.
Neural data #
Neural data streams #
Neural data is collected with Behnke-Fried hybrid electrodes, thus we have 2 neural data streams: micro and macro.
Micro data #
Micro data are recorded on high impedance micro wires at a high sample rate. There are 9 wires in each bundle: one is a low impedance reference wire, the other 8 record data. Micro wires are capable of recording action potentials. We do not record the 9th wire separately, so all 8 channels are permanently bipolar referenced to the 9th.
Macro data #
Aka “intracranial EEG”. These signals are recorded on the large, lower impedance macro contacts of the Behnke-Fried hybrid electrodes. There can be anywhere from 5-12 macro channels per electrode, depending on the length of the electrode chosen for clinical purposes. Action potentials cannot be resolved in these signal.
Data recording systems and sample rates #
We currently have 2 recording systems: Neuralynx (NLX) and BlackRock (BR).
Most (if not all) of the movieParadigm data were recorded with NLX.
Our current data pipeline extracts NLX and BR data with the same folder and filename structure. Due to inherited processing from the previous pipeline, the contents of the files are slightly different (these differences will be included in relevant sections below).
NLX sample rates #
Micro fs = 32000 Hz (32 kHz)
Macro fs = 2000 Hz (2 kHz)
BR sample rates #
Micro fs = 30000 Hz (30 kHz)
Macro fs = 2000 Hz (2 kHz)
NOTE: the nominal sample rate values above are approximate- our recording equipment will have an actual sample rate very close to, but not exactly, the values given above. This is why it is recommended to use the extracted timestamps for analysis, or interpolating to a uniformly sampled time vector if interested in Fourier based signal processing analyses.
Unpacked .mat files #
The first step in our data extraction pipeline is to “unpack” or “extract” our neural data from the raw data files.
The unpacked micro and macro data are stored as an array of electrical time series data. There is no processing done on these data, they are just direct copy of the raw data.
Unpacked data are saved as int16 data types. Within the data file there is a conversion value (ADBitVolts for NLX, BlackRockUnits for BR) to convert the data to a unit of voltage (NLX-> volts, BR -> micro volts).
NOTE: Xin is working on a python function to read the extracted neural data (either from NLX or BR) and load in a consistent manner i.e. with the same units etc.
Data file and folder structure #
The format of the unpacked data follows the structure of the raw data.
Experiments are saved into different folders.

Different data streams are saved into different folders.

Each recording in an experiment is extracted and saved separately.

Filename format #
The brain area at the tip of the planned electrode trajectory is used to “name” the electrodes and resulting data file names from a given electrode. Since the microwires are at the tip of the hybrid electrode these can be considered the microwire targets. However it is important to note that these are targets and not verified recording locations (we do have an imaging pipeline to localize microwire placement but that is not integrated into the file naming convention).
Unpacked micro data are saved with the following file format:
{rec_bank}-{target_area}{channel_number}_{rec_number}.mat
You can generally just ignore the rec_bank information.
Unpacked macro data are saved with the following file format:
{target_area}{channel_number}_{rec_number}.mat
target_area typically follows the format hemisphere-area, or hemisphere-position-area er RAH (right anterior hippocampus), LA (left amygdala). Sometimes the “position” is in lowercase. In some instances there is a hyphen in the area name.
In some rare cases, two bundles have the same target area. These cases have a “-b” at the end of the target_area. Eg 1717 RaiI and RaiI-b
lfpTimestamps files #
Each recording has an associated timestamps file. For every data point there is an associated time value. These timestamps are in seconds, and are in Unix time.
There are also lfpTimeStamps_00X.json files that contain the values of the first and last timestamps in Unix time, for ease of accessing these times for alignment purposes.
Multiple recordings in a data folder and potential time gaps #
There are 2 reasons there can be multiple recordings in one NLX data folder:
- the recording was over 2 hours long. NLX creates a new recording file every 2 hours.
- the NLX recording was paused, but the acquisition was left on (see Schematic above).
In the first case, the timestamps between the 2 recordings should be contiguous ie the time gap between them should be 1/fs.
In the second case, there will be a “large” time gap between the recordings that typically range from seconds to minutes long. This time gap is reflected in the timestamps as they are in the Unix global time frame.
This is not an issue with BR data as the raw data is saved in a different format to the NLX.
Loading unpacked data in python #
Use mat73.loadmat as the unpacked data files are saved as v7.3 in MATLAB.
Handling missing data #
On rare occasions during data acquisition data packets are lost or not fully filled. In these instances the missing sections within the data array are filled with -32,768 (the lowest value of an int16). This has not yet (2024-10-30) been fully implemented across all our data but will be in the near future.
Micro data: spikes #
Our spike data is collected using microwire bundles in a Behnke-Fried hybrid electrode. Fried splays the wires in the bundle so wire tips could be up to 1mm apart. This means that typically action potentials are not recorded across multiple channels.
Spike data filename format #
Manually curated spike data is saved with the following file format:
times_manual_{rec_bank}-{target_area}{channel_number}.mat
Channel numbers range from 1 to 8. You can ignore the rec_bank information. As detailed above the target_area does not reflect actual verified recording locations, just the planned target region.
Spike data format #
Spike detection is run on the full experimental series at the same time to allow us to (try to) track the same units over all experimental epochs.
Spike data is thus saved in folders with names containing the experiment numbers of the full series.

Spike times and cluster identity are contained in a variable named cluster_class.
Cluster_class is a n_spikes by 2 (rows x cols) array. Each row is a spike. The first column contains cluster ID and the second column contains the spike time.
The spike times are in seconds relative to the first timestamp of the first recording of the first experiment in the series (“series reference time frame”; see above).
In each times_manual_*.mat file there is a variable timestampsStart which is the Unix time of the series start. This value can be used to convert times in the single experiment folders to be in the series time frame, and thus aligned with the spiking data. This value can be found in “CSC_micro_spikes_meta.json” files.
Loading spike data in python #
The times_manual_*.mat should all now be saved as v7.3. You should therefore just be able to use mat73 to load them, or you can use a try-except with mat73.loadmat and scipy.io.loadmat to catch any files that may not have been converted:
try:
chan_data = mat73.loadmat(file_path)
except (NotImplementedError, TypeError):
chan_data = scipy.io.loadmat(file_path)
One difference in these loading functions is how they handle the timestampsStart variable:
ts_start = chan_data[“timestampsStart”] # if loaded with mat73 this will extract value
if ts_start.shape == ():
ts_start = float(ts_start)
else:
ts_start = float(ts_start[0][0]) # if loaded with scipy have to extract value from array
Micro data: LFP #
We are working on processing the micro data to extract the micro LFP signal which will have action potentials removed and be downsampled to 2kHz. Analyzing micro LFP without removing spikes may lead to bleedthrough into high frequency (50-200 Hz) channels.
Auxiliary data streams #
TTL/events #
TTLs are extracted for each experiment.

TTLs.csv contain every TTL sent during the experiment that could be matched in both the neural DAQ and the matlab logs.

TTL_table.csv contains a restructured version of the TTLs into epochs or trials

Later sessions will contain slightly more information following changes to the task code but the timing information in both reflect the same points in the task.

The DAQ timestamps given are in Unix time so are in the same time frame as the extracted timestamps arrays, meaning they can be used to align behavioral events to the neural data streams.
Note on recall data: The recall TTL information indicates the start and stop time of the 5 minutes given for recall. Participants are asked to continue if they can (later TTL files contain information as to when the participants ‘click to continue’ after finishing recall). A number of participants speak for less than 5 minutes. This is to say for the most accurate timing available on when a participant is recalling it is recommended to use the audio alignment.
More details on the TTL output will be added when Soraya has time.
Important note on TTL accuracy #
For all recordings before Oct 2024 there is a 30 ms delay following each TTL sent. From Oct 2024 onwards this delay will be 1ms.
We are also currently (2024-10-25) investigating a potential ~150 ms delay from the TTL movie start times to the actual movie onset as measured by audio-alignment. So for movie onset times we would currently advise using the audio-aligned start times (found on the Fried Lab Movie task notes–ask John or Soraya for a copy) where possible. We believe this is an operating system issue involving psychtoolbox video presentation on mac/windows. We are investigating the impact of this issue on the TTLs during the recognition memory where short video clips are presented.
Audio #
NOTE: The full details of the audio data extraction can (probably) be found elsewhere. The information here is just primarily focusing on timing and alignment.
Audio recordings are saved with the DAQ and typically also with a backup device (eg laptop/phone).
Audio-aligned movie start times #
John has been finding the time offset between experiment start time and movie start time by correlating the DAQ recorded audio and the audio stream of the presented video files.
- times given are all in the experiment reference frame.
- Ask John or Soraya for a copy of the Fried Lab Movie task notes where we provide the offset between the neural recording and the movie start
For recent patients (565 onwards) the movie start time can also be found in the TTLs.
IMPORTANT NOTE: as described above we are investigating a potential ~150 ms delay from the TTL movie start times to the actual movie onset. Currently we would advise to use the audio-aligned start times where possible (on the Fried Lab Movie task notes) since those are right within ±5 ms.
For a number of control video sessions we do not have TTL info so the movie start time will need to be found using audio alignment. This is a WIP.
The movie start time in Unix can be found in *_movie_start_time.json files in the individual experiment folders, in a folder named “Audio”:

These jsons were generated using information from the previous data extraction pipeline so contain information about that process.
You only need the field start_unix to align the movie time and the spikes:
start_unix minus timestampsStart (from the times files or CSC_micro_spikes_meta.json) = the movie start time in the same reference frame as the spike data
For reference, the *_movie_start_time.json files also contain:
"pID": participant ID "exp_num": experiment number "rec_num": the recording number within the experiment that contains this movie watch "raw_data_folder": the raw DAQ data folder "audio_filename": the audio file from which the movie audio was extracted "gsheet_start": the movie start time (as recorded on a google sheet from a previous data pipeline) “gsheet_pause”: times the movie was paused "start_rel_rec": the movie start time in seconds with respect to the start of the specific recording it is in "exp_t0_unix": the Unix start time of the experiment in seconds "rec_t0_unix": the Unix start time of the recording that the movie watch is in, in seconds "time_shift_applied": the shift in seconds applied to the google sheet times to find the correct times that align with the neural data "start_unix": the Unix time of the movie start "pause_unix": the Unix time of the movie pauses "time_units": the units for all times given in the json -> "seconds" "json_created": the date and time that the json was generated eg "2024-10-18 12:38:13" "note": a description of the processing that occurred to generate the Unix times
Free recall audio #
Quick summary of current pipeline:
- DAQ audio is extracted by Anthony who listens and notes the start and stop times of free and cued recall.
- These times are saved on the ‘Fried Lab Movie task note’ google sheet.
- These times are used to extract cropped audio files which contain only the recall.
- This cropped audio file is what is used for PTR annotations.
- The word onset times found in PTR are relative to the start of the cropped audio file.
- To align the word onset times to the neural data you need to use the start time of the cropped audio file.
- There are 2 cases in which the times in the google sheet will not give an an accurate offset to align the word onset times with the neural data:
-
- If the filename mentions ‘back up’ then the audio files used were not recorded with the DAQ. The times in the sheet are relative to the start of the back-up audio file and not aligned with the DAQ recording. This is the case for 570 (see more details below).
- Pausing NLX during recording while leaving acquisition on (see schematic above) results in multiple recordings in one experiment folder separated by a gap in time. If recall occurred in recording 2+, the concatenation used in the previous pipeline means that gaps between recordings are not accounted for in the timing of recall start and stop recorded on the google sheet.
As all the timing in the individual experiment folders are in the global Unix time frame, the recall start and stop times have been converted to Unix. Any time gaps in current data (prior to 2024-10-18, participants <=577) have been accounted for during the conversion to Unix.
The unix times of the recall start and stop times can be found in json files in the ‘Audio’ folder in each Experiment folder. The file names are the raw audio filenames with the suffix “_recall_timing.json” added.
Eg:

The _timing.json files contain:
"pID": participant ID "exp_num": experiment number "rec_num": the recording number within the experiment that contains this recall "raw_data_folder": the raw DAQ data folder "audio_filename": the audio file from which the recall audio was extracted "gsheet_start_str": the HH:MM:SS time of recall start that is recorded on the google sheet "gsheet_end_str": the HH:MM:SS time of recall end that is recorded on the google sheet "gsheet_start": gsheet_start_str converted to seconds "gsheet_end": gsheet_end_str converted to seconds "start_rel_rec": the recall start time in seconds with respect to the start of the specific recording "end_rel_rec": the recall end time in seconds with respect to the start of the specific recording "exp_t0_unix": the Unix start time of the experiment in seconds "rec_t0_unix": the Unix start time of the recording that free recall is in, in seconds "time_shift_applied": the shift in seconds applied to the google sheet times to find the correct times thatalign with the neural data "start_unix": the Unix time of the recall start "end_unix": the Unix time of the recall end "time_units": the units for all times given in the json -> "seconds" "Json_created": the date and time that the json was generated eg "2024-10-18 12:38:13" "note": a description of the processing that occurred to generate the new Unix times
To align the word onset times from PTR with the neural data you just need to add the start_unix value to the word onset times.
NOTE: For participants before 2024-10-18 recall start and stop times were recorded on the google sheet, and the conversion to Unix was processed later. For patients after 2024-10-18 the timing json files are made by Anthony’s audio pipeline – this will be indicated in the “note” section.
Video drift #
Any neural recording requires alignment to the behavior. When presenting short stimuli experimenters typically use TTLs sent from the behavioral computer to the neural computer to record the time of stimulus presentation in a channel recorded along with the neural data. This is because even if you know the offset from the beginning of the neural recording to the beginning of the behavioral experiment, the two computers running each will slowly drift away from each other due to the sub-millisecond imprecision of quartz clocks. By re-locking the stimuli on every presentation (typically a few seconds apart) you don’t have to worry about this drift.
Unfortunately, for videos, we have come to realize that whether playing it using VLC or within PsychToolBox, the behavioral computer fails to match the intended FPS for our videos. For example, the 24 S06E01 .m4v file is meant to be shown at 29.97 FPS. Unfortunately, different laptops (unintentionally) play the video at slightly slower or faster FPS, which creates a linear drift away from the “ground truth” expectation you’d have while analyzing movie frames (since your analysis software assumes the movie was shown at precisely 29.97 FPS). This typically leads to the neural data being misaligned from what you’d expect by 10-200 ms over the course of a video’s presentation.
To account for this drift, John has written software that aligns the audio recorded during patient video viewing with the movie file. Essentially, it reads in the movie file and the recorded audio from a given session, resamples them to the same samples/time, and aligns the audio between them to find the offset between the neural recording and the video start at various points throughout. Here’s an example of the output plot:
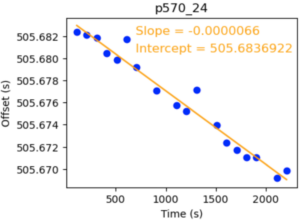
Despite some variability, the 18 points sampled throughout the movie provide a linear fit that lets us know the timing of what the patient saw relative to how far into the movie they are (x-axis). Therefore, the intercept gives you the offset between the neural recording start and the movie start at the beginning of the movie, and the slope tells you how much the video drifts over time (in seconds) compared to the ground truth FPS.
With this intercept and slope you can adjust your neural analysis to match what the patient saw. To correct for a negative slope, which means your movie is playing faster than the expected FPS (by playing too fast the movie is ‘catching up’ and therefore reducing the offset in the plot), you need to get the correction factor like this: correction_factor = 1+drift_rate_slope. E.g. for this one that comes to 0.9999934, meaning any event the patient was shown in the movie happened at time T*0.9999934 compared to an FPS of 29.97 to account for the video being played slightly too fast.
The same formula works for a movie playing too slow (with a positive drift rate. E.g. for p577 the drift rate is 9.68E-5). Plugging that into the formula above, you get 1.000968. Which means since the movie was being played too slow (which happens on our Linux laptop) you need to multiply things that happened in the movie by that factor to account for the patient seeing them slightly later in time than expected by the movie’s FPS.
Drift rates calculated for our various video tasks are on the “Fried Lab Movie task notes” google sheet. (Ask Soraya or John if you need access).
Bespoke patient preproc/fixes #
568 macro montage channel name fix #
The NLX montage included an extra ROPR-AI channel – there were 7 when there should have been 6. All channel names after this point were shifted by one. We made a copy of the raw on the LTS and fixed the naming, and then extracted these data to hoffman. So all extracted data should have the correct name. We did not lose any neural data, might have lost polysom A2.
571 Exp5 lost data packets #
There were some lost data packets for a subset of channels in 571 Exp5. The gaps were filled with values of -int16.
570 back-up audio processing #
While aligning the back-up audio to the NLX audio John realized the back-up audio is drifting wrt the NLX audio. He has found the offset between the NLX audio and back-up audio at the specific times that the recall sections were cut from the full back-up audio file to account for any drift that occurred before that point.
The above timing offsets have been added in the timing json files for 570, so the “start_unix” value can be used to align the PTR word onset times like all the others.
1789 Exp101 (sleep) empty files #
The files uploaded from Iowa for PDes88_0002.ncs and PDes88_0003.ncs were zero bytes. Asked Iowa and they think perhaps it was an upload error from their NLX cart as they were also empty on their server. Unfortunately these files were deleted from the NLX cart so cannot be recovered.
PDes88 maps to LSMG8. To be able to run spike sorting without modifying the code I have made two placeholder files (LSMG8_003.mat, LSMG8_004.mat) with all the same metadata as the other files, but now data is a vector of zeros (the same length as the other _003/_004 recordings).
The code has been edited to exclude these from the median calculated for CAR.
This also means that we will only have any spikes for the first 4 hours of the sleep rec for this channel. For all other experiments the data for this channel appears to be present.

